-Search query
-Search result
Showing 1 - 50 of 333 items for (author: shanshan & l)

EMDB-42820: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5b extended
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-35595: 
Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex
Method: single particle / : Zhang X, Duan ZQ, Zhu JK

PDB-8inb: 
Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex
Method: single particle / : Zhang X, Duan ZQ, Zhu JK

PDB-8j5y: 
Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery
Method: single particle / : Chen J, Chen H, Li S, Lin X, Hu R, Zhang K, Liu L

PDB-8j60: 
Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery
Method: single particle / : Chen J, Chen H, Li S, Lin X, Hu R, Zhang K, Liu L

EMDB-42816: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyp: 
SARS-CoV-1 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42801: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42802: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42805: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42808: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 4
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42809: 
MERS 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42810: 
MERS 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42811: 
MERS 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42818: 
SARS-CoV-2 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42819: 
SARS-CoV-2 5' proximal stem-loop 5 and 6 with SL5c extended
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42821: 
SARS-CoV-2 5' proximal stem-loop 5 and 6
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uye: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyg: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyj: 
BtCoV-HKU5 5' proximal stem-loop 5, conformation 4
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyk: 
MERS 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uyl: 
MERS 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uym: 
MERS 5' proximal stem-loop 5, conformation 3
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

PDB-8uys: 
SARS-CoV-2 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42803: 
HCoV-229E 5' proximal stem-loop 5
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42813: 
HCoV-NL63 5' proximal stem-loop 5, conformation 1
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-42814: 
HCoV-NL63 5' proximal stem-loop 5, conformation 2
Method: single particle / : Kretsch RC, Xu L, Zheludev IN, Zhou X, Huang R, Nye G, Li S, Zhang K, Chiu W, Das R

EMDB-34000: 
Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34001: 
Spiral hexamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34002: 
Spiral pentamer of the substrate-free Lon protease with a S678A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34003: 
Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34004: 
Spiral hexamer of the substrate-free Lon protease with an S678A mutation
Method: single particle / : Li S, Hsieh KY, Kou CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34005: 
Open-spiral pentamer of the substrate-free Lon protease with Y397A and S678A mutations
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34006: 
Spiral hexamer of the substrate-free Lon protease with Y397A and S678A mutations
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34107: 
MtaLon-Apo for the spiral oligomers of trimer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI
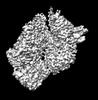
EMDB-34108: 
MtaLon-Apo for the spiral oligomers of tetramer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI

EMDB-34109: 
MtaLon-Apo for the spiral oligomers of pentamer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI

EMDB-34110: 
MtaLon-Apo for the spiral oligomers of hexamer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI
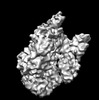
EMDB-34111: 
MtaLon-ADP for the spiral oligomers of trimer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI
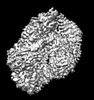
EMDB-34112: 
MtaLon-ADP for the spiral oligomers of tetramer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI

EMDB-34113: 
MtaLon-ADP for the spiral oligomers of pentamer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI

EMDB-34114: 
MtaLon-ADP for the spiral oligomers of hexamer
Method: single particle / : Li S, Hsieh K, Kuo C, Lee S, Ho M, Wang C, Zhang K, Chang CI

EMDB-34116: 
Spiral pentamer of the substrate-free Lon protease with an M217A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-34117: 
Spiral hexamer of the substrate-free Lon protease with an M217A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

EMDB-36865: 
Spiral pentameric form of the substrate-free Lon protease with a Y224S mutation in the presence of the N-terminal-truncated monomeric mutant bearing an E613K mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Zhang K, Chang CI

EMDB-36866: 
Spiral hexameric form of the substrate-free Lon protease with a Y224S mutation in the presence of the N-terminal-truncated monomeric mutant bearing an E613K mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Zhang K, Chang CI

EMDB-36867: 
The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant
Method: single particle / : Li S, Hsieh KY, Kuo CI, Zhang K, Chang CI

PDB-7yph: 
Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

PDB-7ypi: 
Spiral hexamer of the substrate-free Lon protease with a Y224S mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI

PDB-7ypj: 
Spiral pentamer of the substrate-free Lon protease with a S678A mutation
Method: single particle / : Li S, Hsieh KY, Kuo CI, Lee SH, Ho MR, Wang CH, Zhang K, Chang CI
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
